Here, we present a list of projects developed and conducted by members of ISyN. These projects showcase analysis pipelines and concepts, including codes, to be downloaded and used by ISyN members. You are encouraged to share your project, for that, please contact isyn@lir-mainz.de.
Development of tools for dynamic systems analysis in resilience research (IDSAIR) in the context of a cooperation project between the Leibniz Institute of Resilience Research in Mainz and the Institute of Industrial Mathematics (Fraunhofer ITWM) in Kaiserslautern, funded by the state of Rhineland-Palatine.
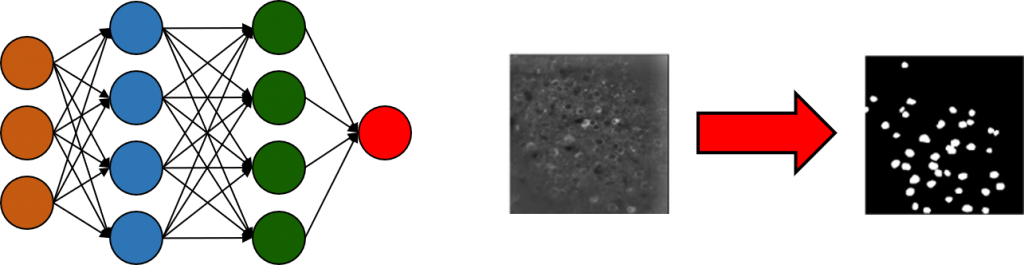
IDSAIR Project I – Optimization of the Calcium Imaging Analysis Pipeline
Calcium Imaging Analysis enables researchers to track the activity of hundreds and thousands of neurons within the brains of living animals. However, analyzing this huge amount of data raises some difficulties. In order to improve and accelerate the analysis of calcium imaging data, we collaborate with the Fraunhofer ITWM and the group of Prof. Dr. Albrecht Stroh (http://www.strohlab.com/) to automatically label neurons in the produced image files using Deep Learning. Following this part of the analysis, the peaks within the transients of the identified neurons must be identified. By optimizing the whole analysis pipeline and making the procedure available and easily useable for other researchers, the effort and time spent with such analyses can be drastically reduced. Furthermore, results will be better comparable as the analysis will be less dependent on personal decisions.
IDSAIR Project II – Optimization of Standard Bioinformatic Pipelines
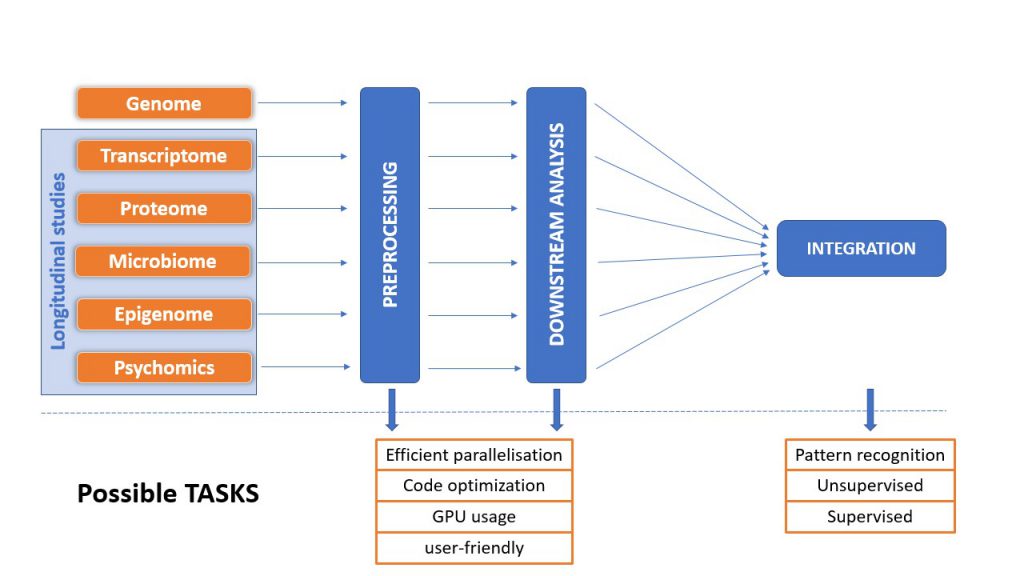
Typical bioinformatics pipelines for analyzing omics data consist of a variety of tools – written in different programming languages by different institutes. As some of these applications are not constructed to be run in parallel on GPU-based hardware, these application can create a bottleneck in terms of speed when running bioinformatics analyses. In collaboration with the Frauenhofer ITWM we want to optimize the separate tools of typical omics-data analysis pipelines to make them run on GPU based machines, thus accelerating omics-data analyses.
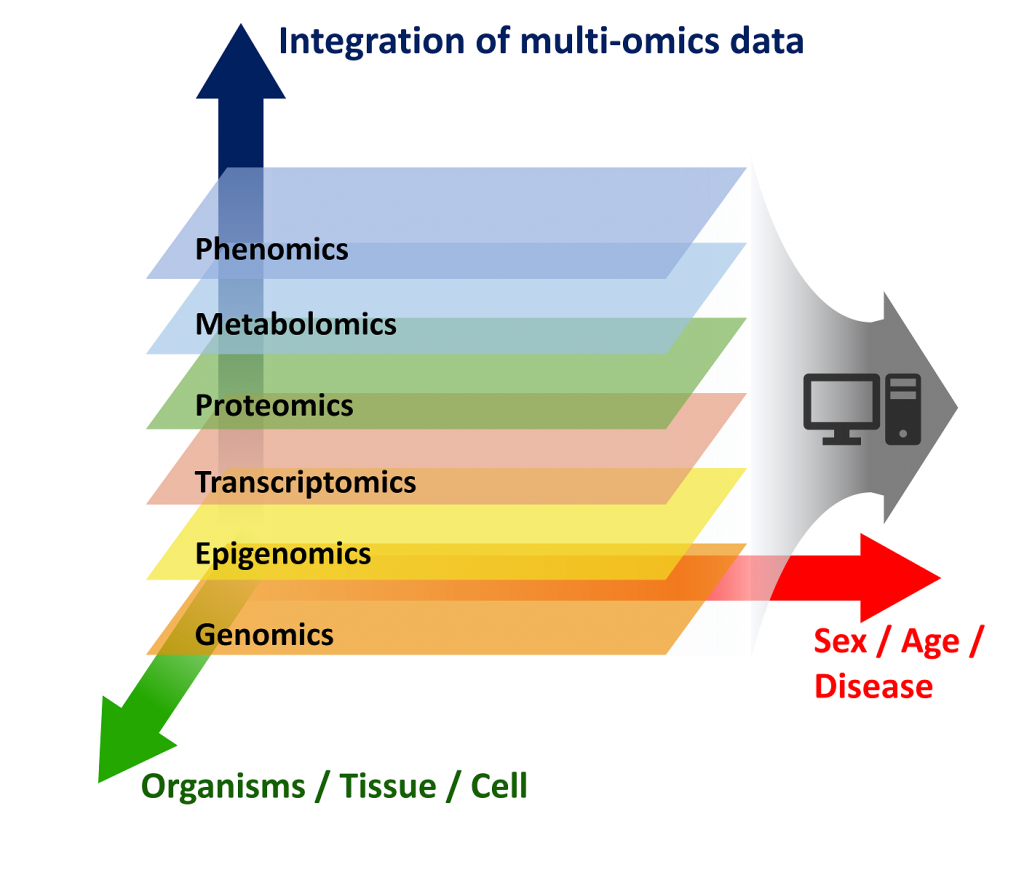
IDSAIR Project III – Using Multi-Omics Integration to explore the Molecular Background of Stress Resilience
Resilience is the ability to cope with stress or to quickly recover to pre-crisis state after being exposed to extreme stress. As the human mental status is highly diverse, there are presumably many different molecular mechanisms underlying resilience. The main goal of this project is the identification of common molecular patterns between resilient individuals by analyzing and integrating various omics levels, e.g. transcriptomics, proteomics, methylomics and metagenomics. By subsequently selecting important features within the large amount of data, we aim to give a less complex view on the dynamic process of resilience.
Collaborations: LIR, Frauenhofer ITWM
Python GUI for analyzing IntelliCage data

The IntelliCage system helps researchers to conduct behavioral experiments and learning experiments with mice while ensuring minimal human intervention. The animals can be observed for long time periods – up to several weeks. This long-term data acquisition can provide new insights in mouse behavior, that might not be detectable in short-term observations.
However, analyzing those big amounts of data is challenging for many researchers.
IntelliPy aims to simplify and automize many aspects of the analysis, such as acquiring data per group, creating learning curves or pivoting parameters in different timeframes. All plots are automatically created and the final tables for statistical tests are stored separately for the user.
The related publication can be found here: https://pubmed.ncbi.nlm.nih.gov/34601559/
Github: https://github.com/NiRuff/IntelliPy
People: Nicolas Ruffini in collaboration with RG Müller (Franziska Mey, Giulia Treccani, Ulrich Schmitt)
StaAv – Stack Averaging Tool
An ImageJ Plugin that can be used to mark and copy frames from an image stack to a new stack to create an average image with enhanced cell visibility from the new stack. This Plugin can be used either by manually selecting only the frames with cell activity, by automatically marking a frame for every half of the calcium indicator decay time or both.
Github: https://github.com/Strohlab/StaAv-Tool
People: Saleh Altahini
Event-related Analysis Pipeline for functional Calcium Imaging Data
The goal of any pipeline for data processing is to extract a subset of features from the raw data, either based on a priori hypotheses, or by data-driven unsupervised methods. Here, we present a pipeline based on the idea of an event-related analysis: We put forward the notion, that any functional circuit imaging of a neuronal ensemble at the end only serves as a tool to extract the underlying action potential train of each individual neuron. Consequently, we provide a step-by-step guide to extract putatively AP-related calcium transients form the raw images. Once this optically derived matrix of AP-related events is generated, e.g. by transforming an extracted intensity trace to a binarized train of zeros and ones, already well-established correlation or interaction concepts can be employed on these dimensionality-reduced datasets.
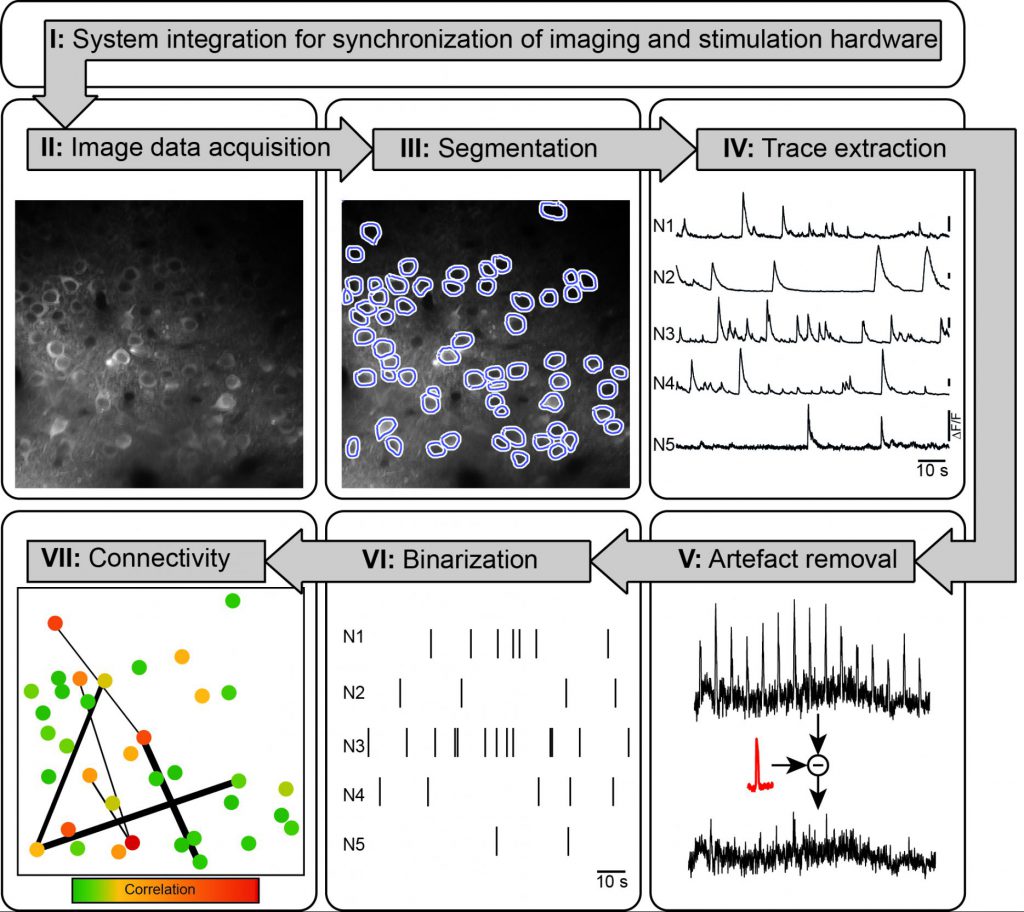
Github: https://github.com/Strohlab
People: Hendrik Backhaus
